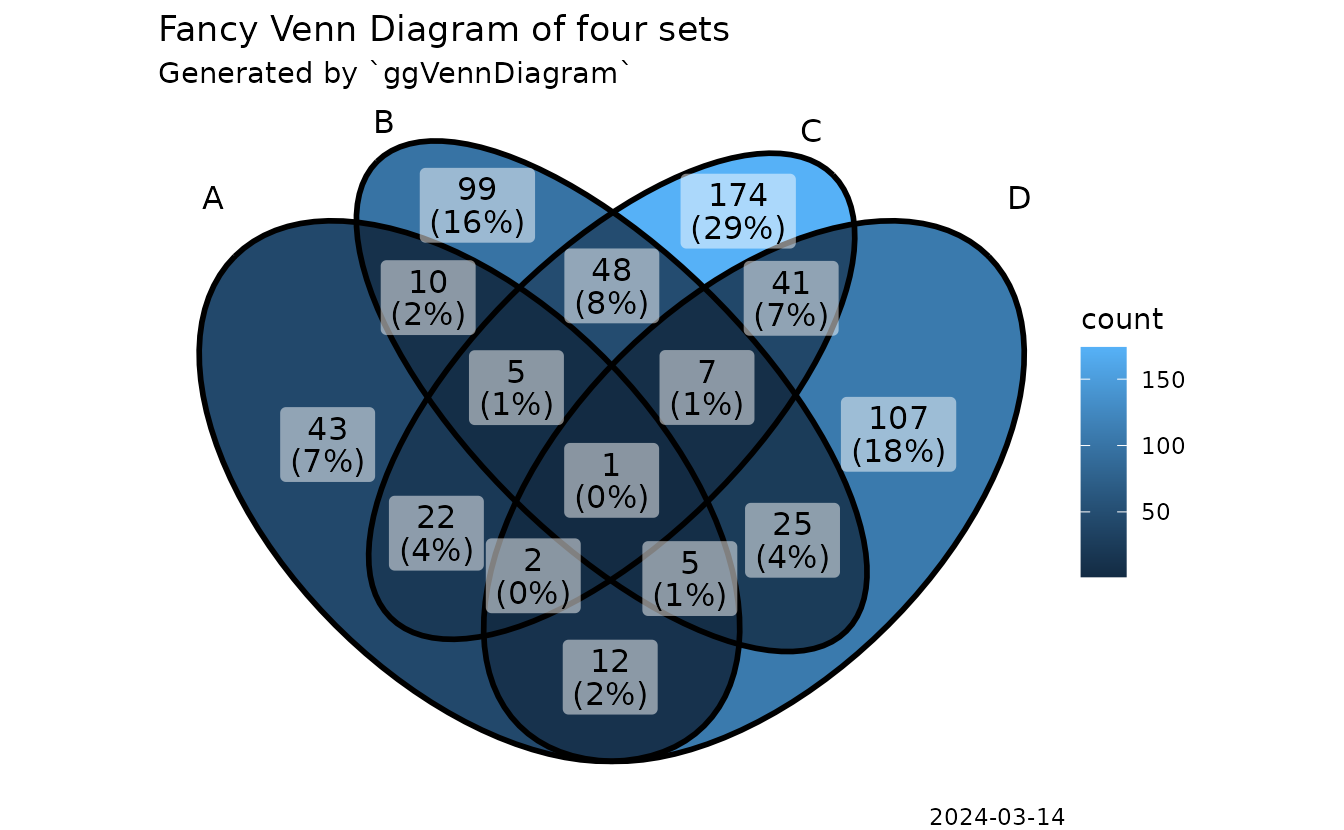
‘ggVennDiagram’ enables fancy Venn plot with 2-7 sets
and generates publication quality figure.
Installation
You can install the released version of ggVennDiagram from CRAN with:
install.packages("ggVennDiagram")And the development version from GitHub with:
# install.packages("devtools")
devtools::install_github("gaospecial/ggVennDiagram")long category names
If you use long category names in Venn plot, labels may be cropped by plot borders. To avoid this, just use a ggplot trick to expand x axis.
p1 <- ggVennDiagram(gene_list,
category.names = c("a very long name","short name","name","another name"))
p1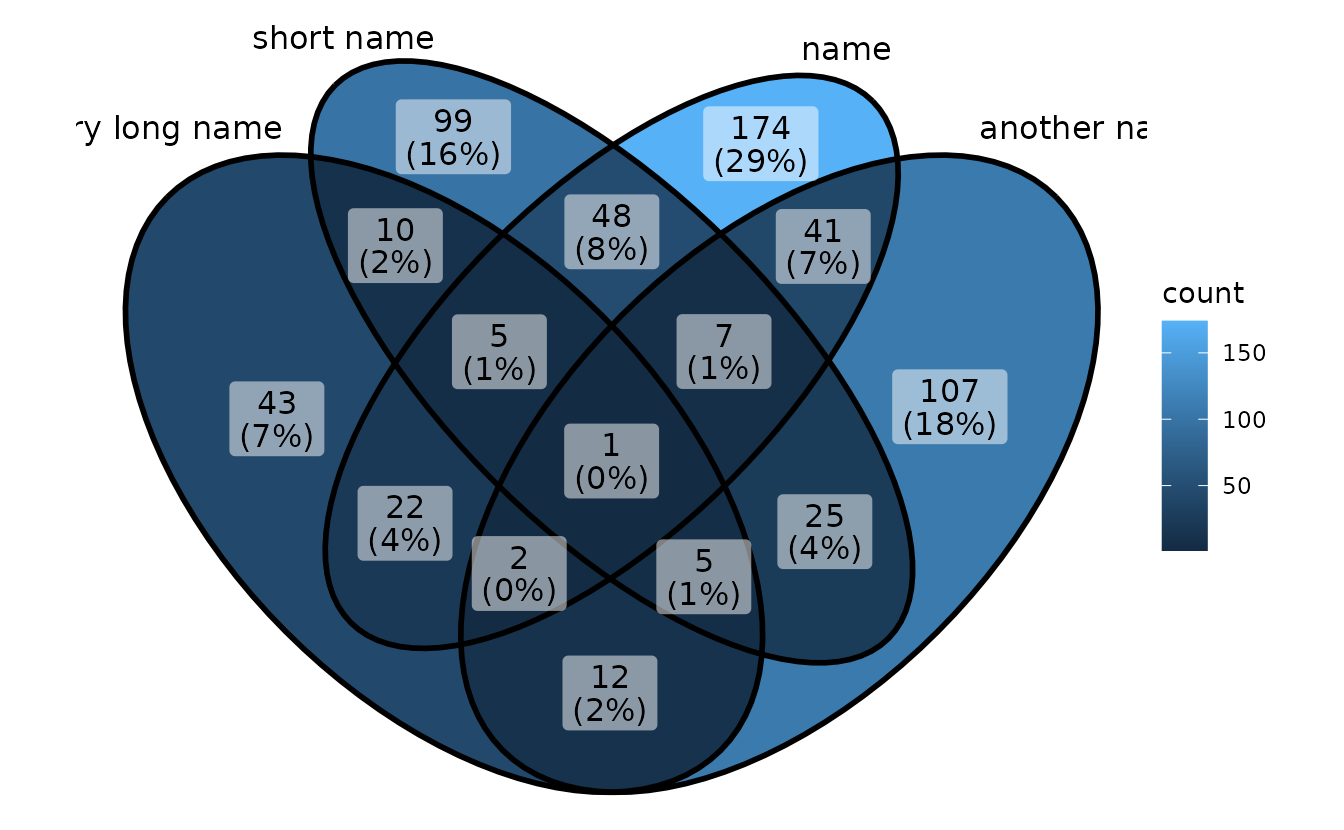
# expand axis to show long set labels
p1 + scale_x_continuous(expand = expansion(mult = .2))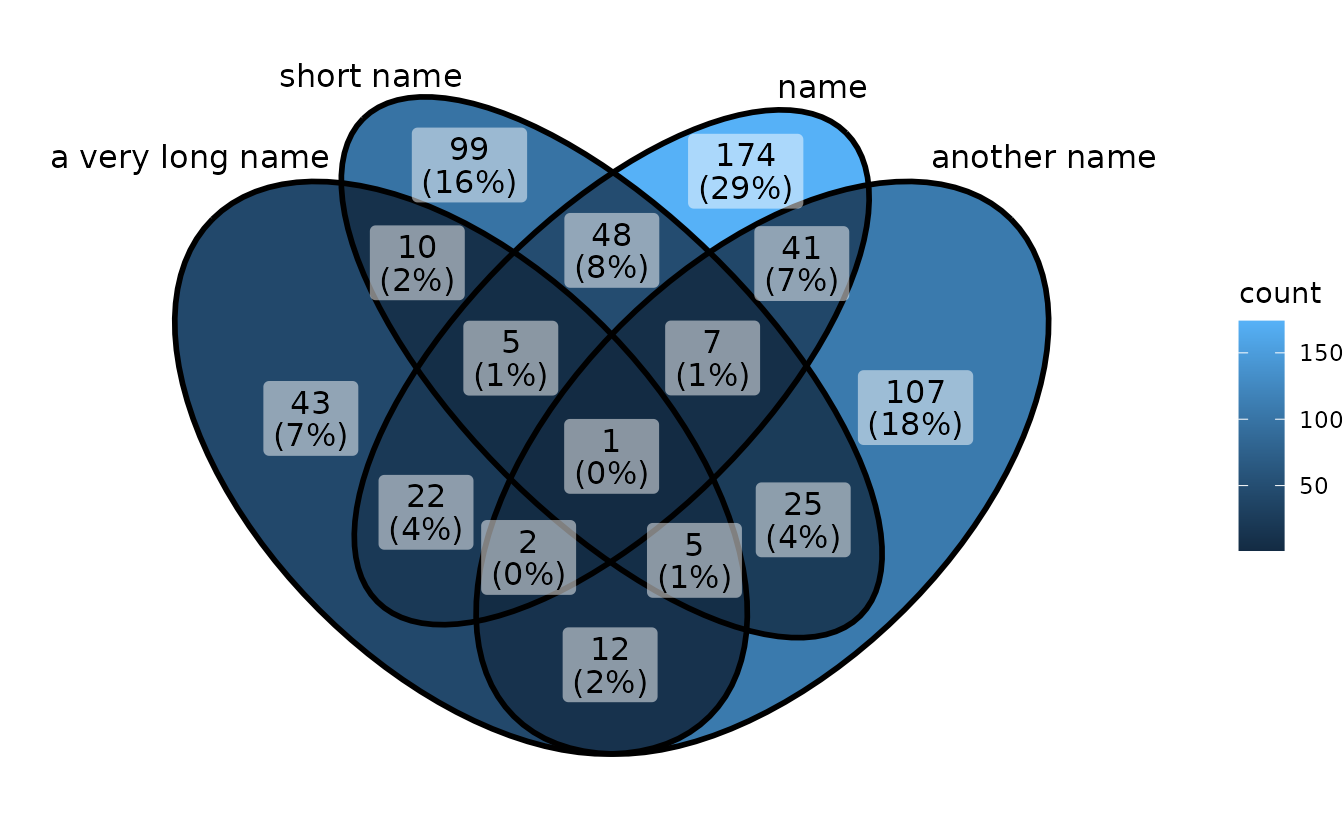
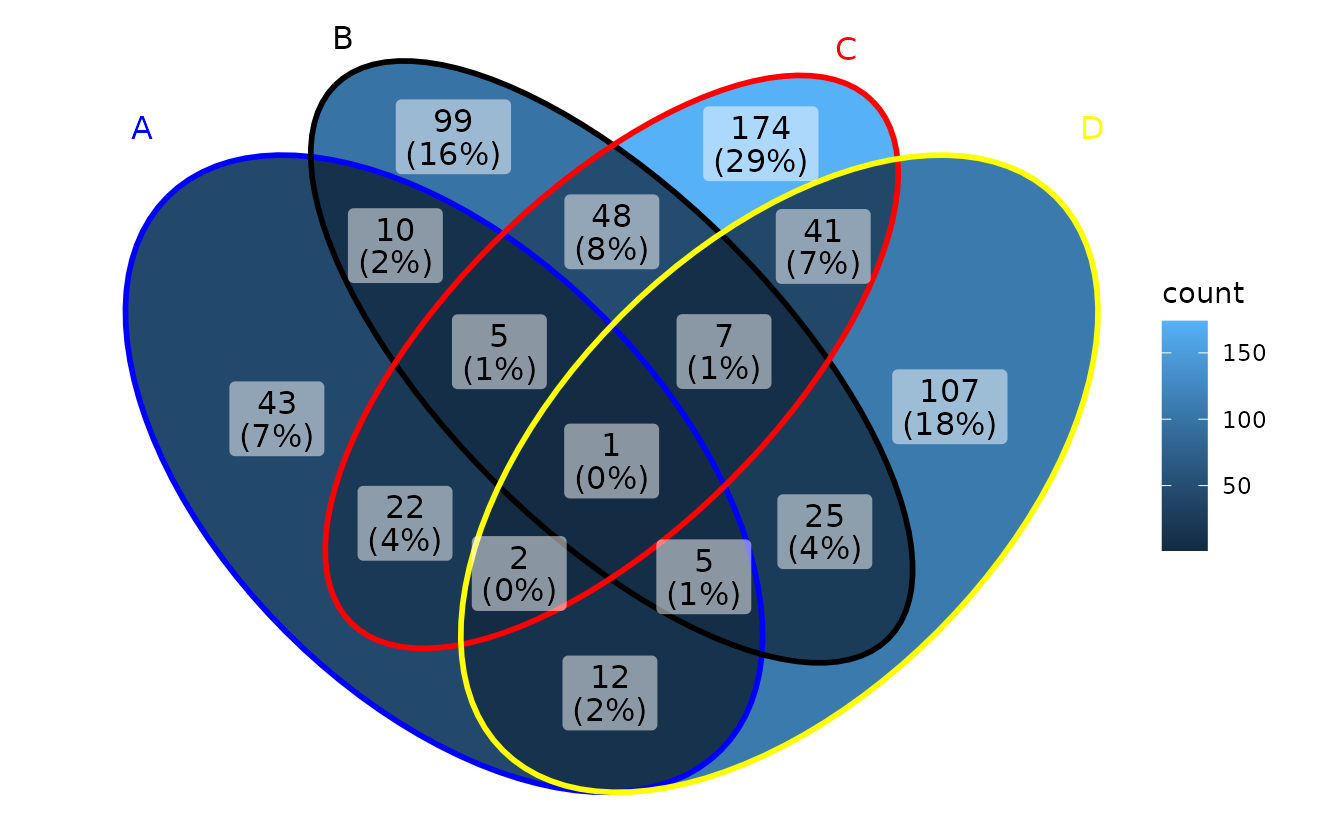
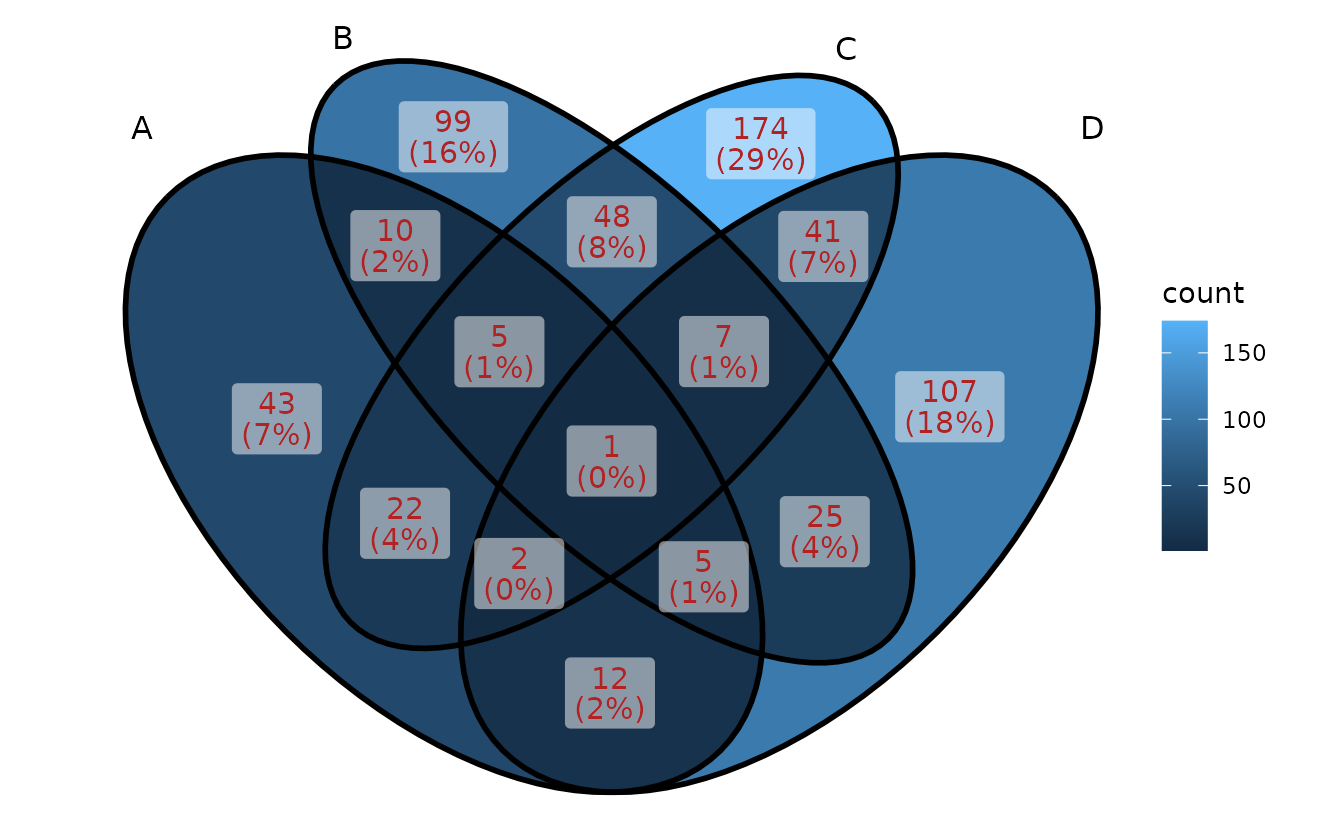
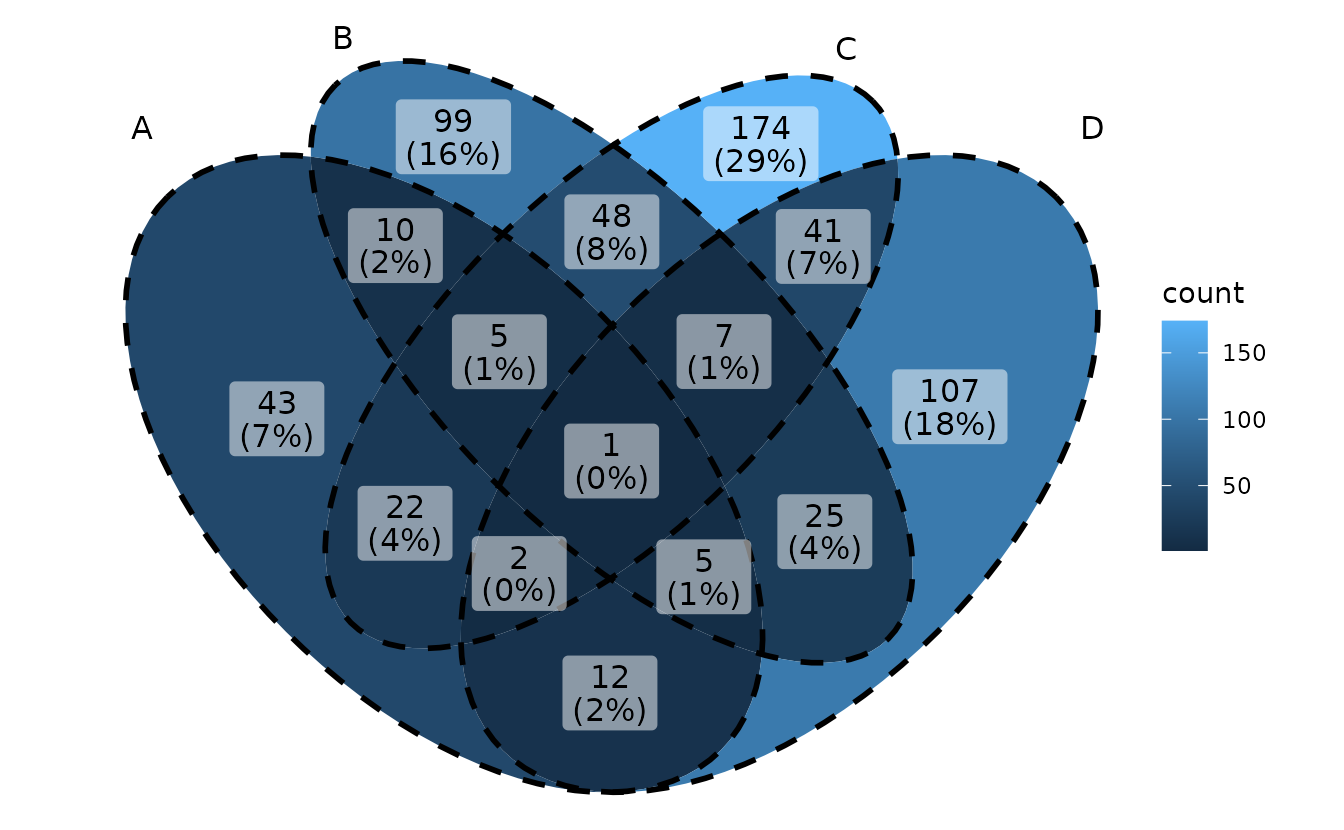
Show intersection values
When intersection values only have several members,
ggVennDiagram is efficient to show the values in
places.
set.seed(0)
small_list <- lapply(sample(6:10, size = 4), function(x){
sample(letters,x)
})
ggVennDiagram(small_list,
category.names = LETTERS[1:4],
show_intersect = TRUE)
#> Warning in geom_text(aes(label = .data$count, text = .data$item), data =
#> region_label): Ignoring unknown aesthetics: textSetting region label
text content
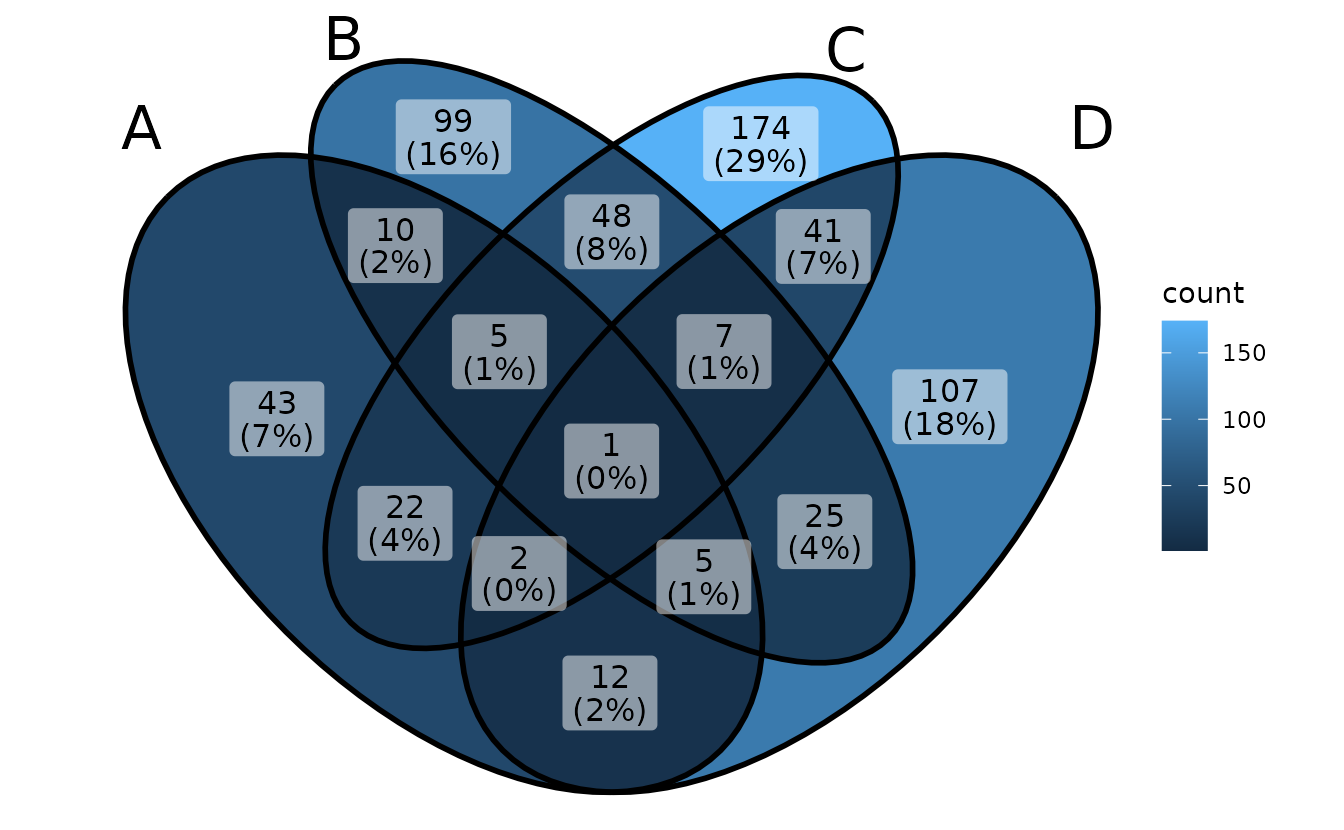
ggVennDiagram(gene_list, label = "count")
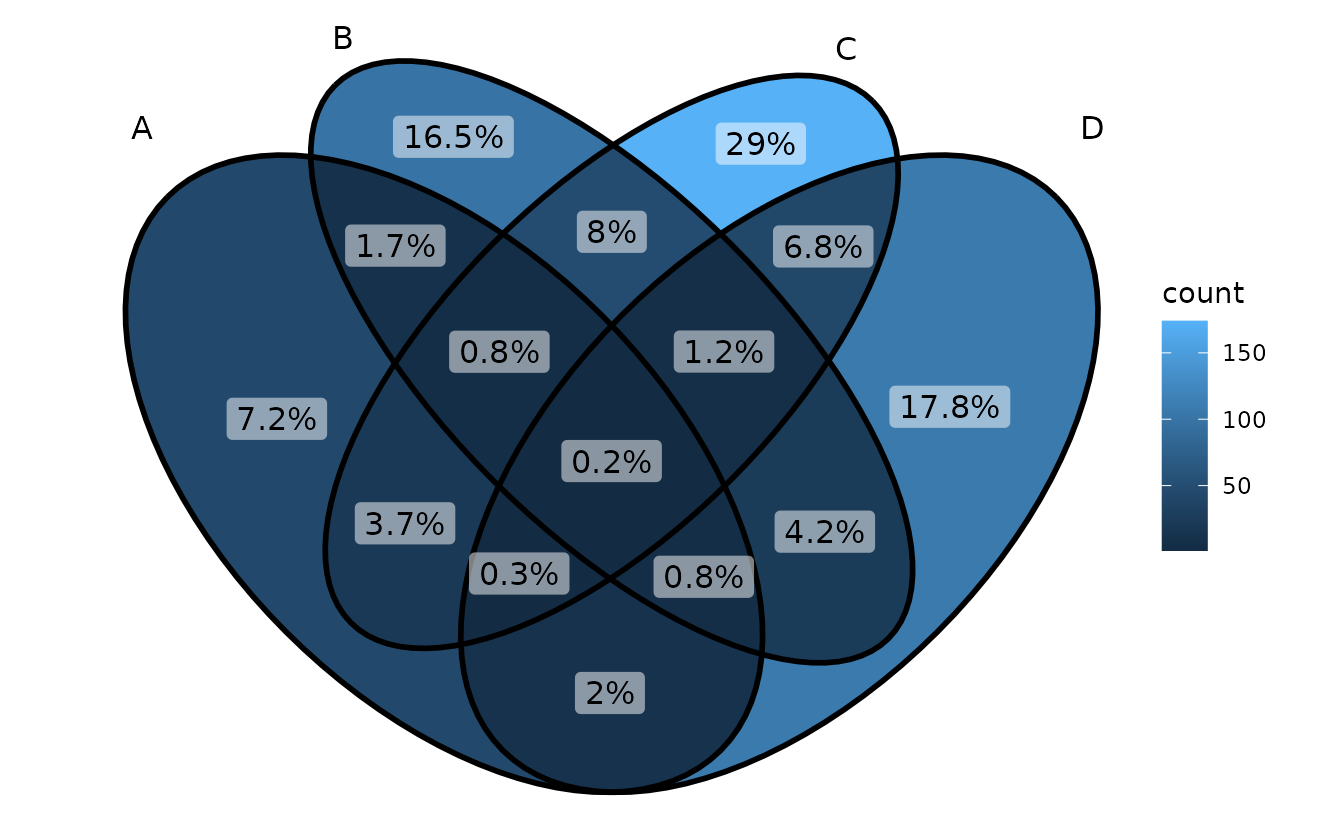
ggVennDiagram(gene_list, label = "percent")
ggVennDiagram(gene_list, label = "both")
ggVennDiagram(gene_list, label = "none")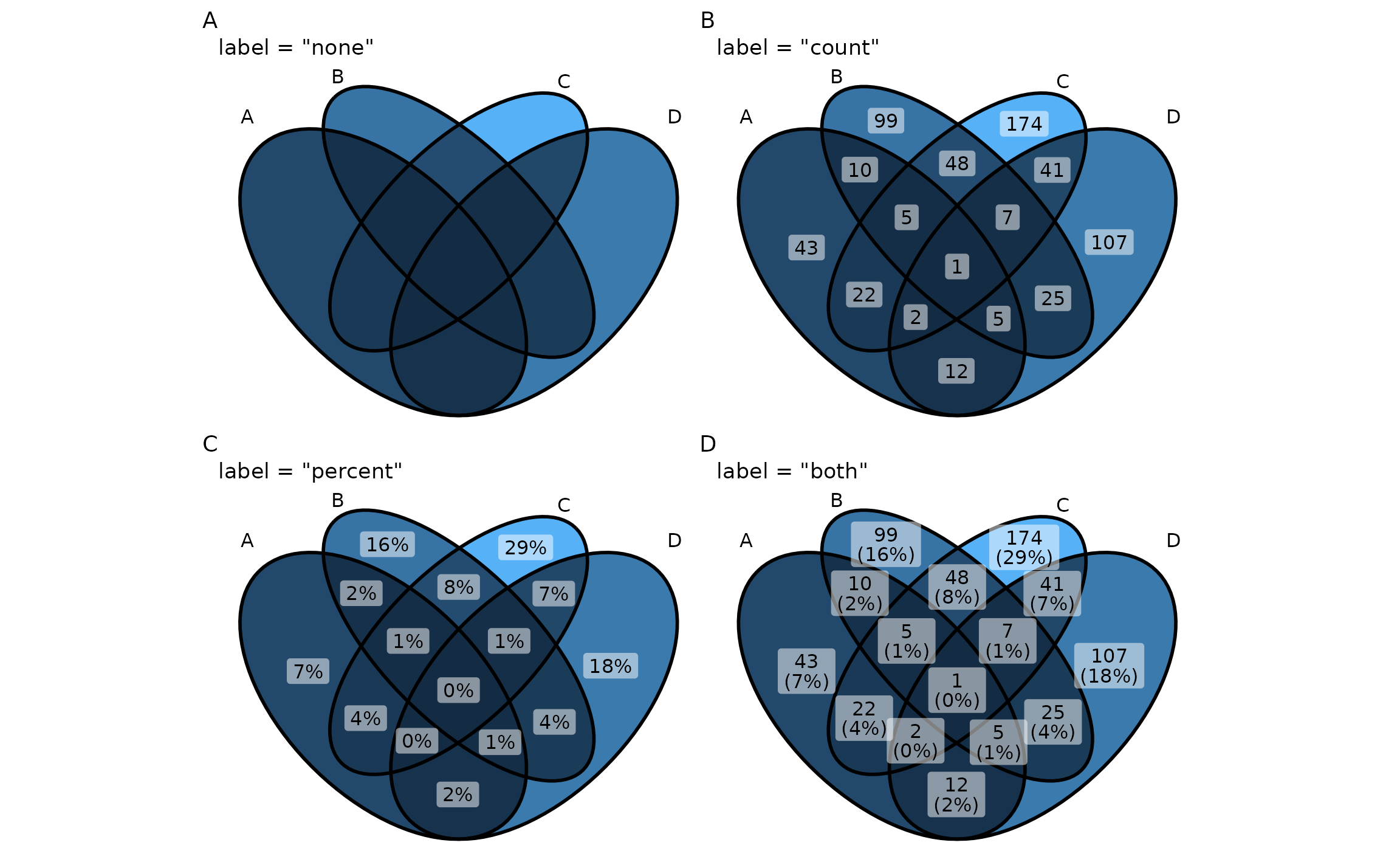
remove label background
- Method 1: set alpha to 0
ggVennDiagram(gene_list, label_alpha = 0)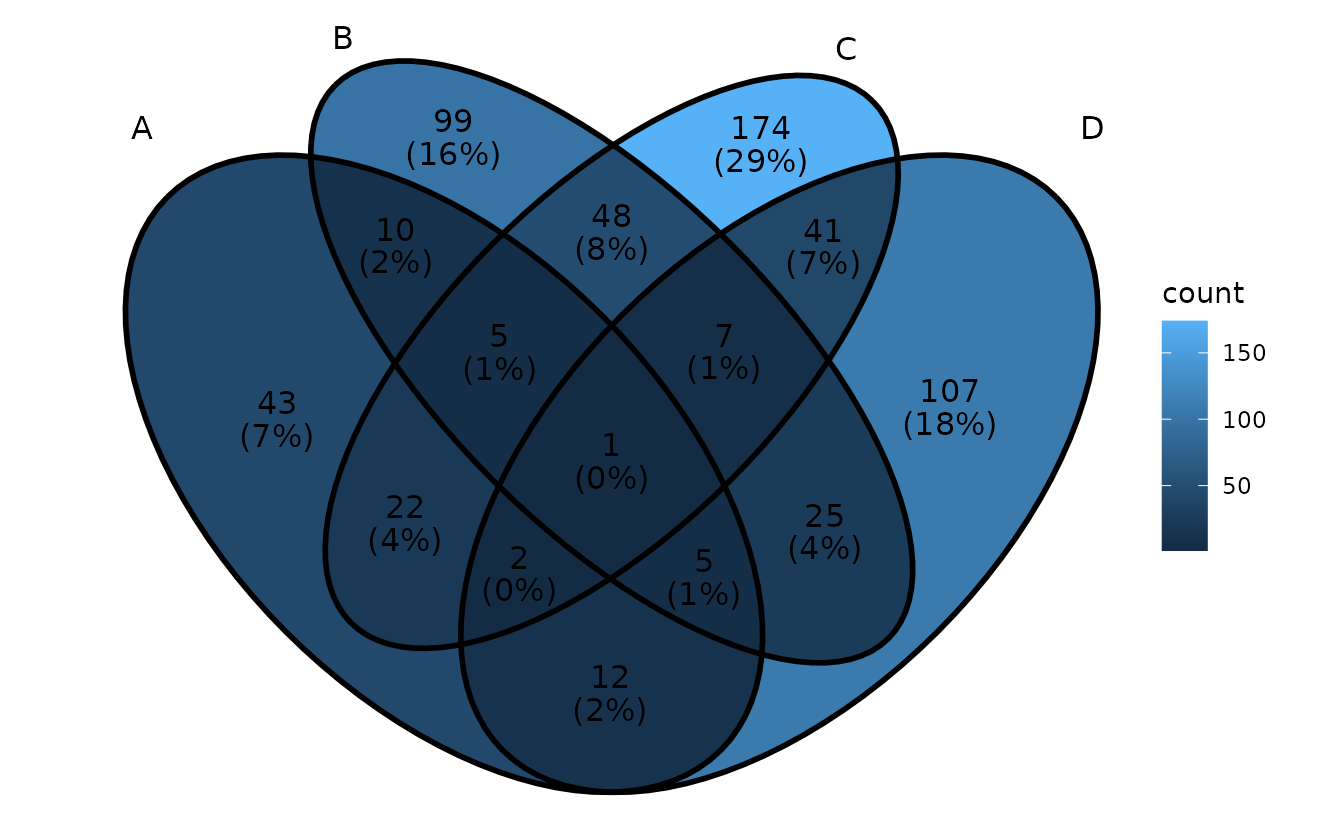
- Method 2: use
geom_text()
ggVennDiagram(gene_list, label_geom = "text")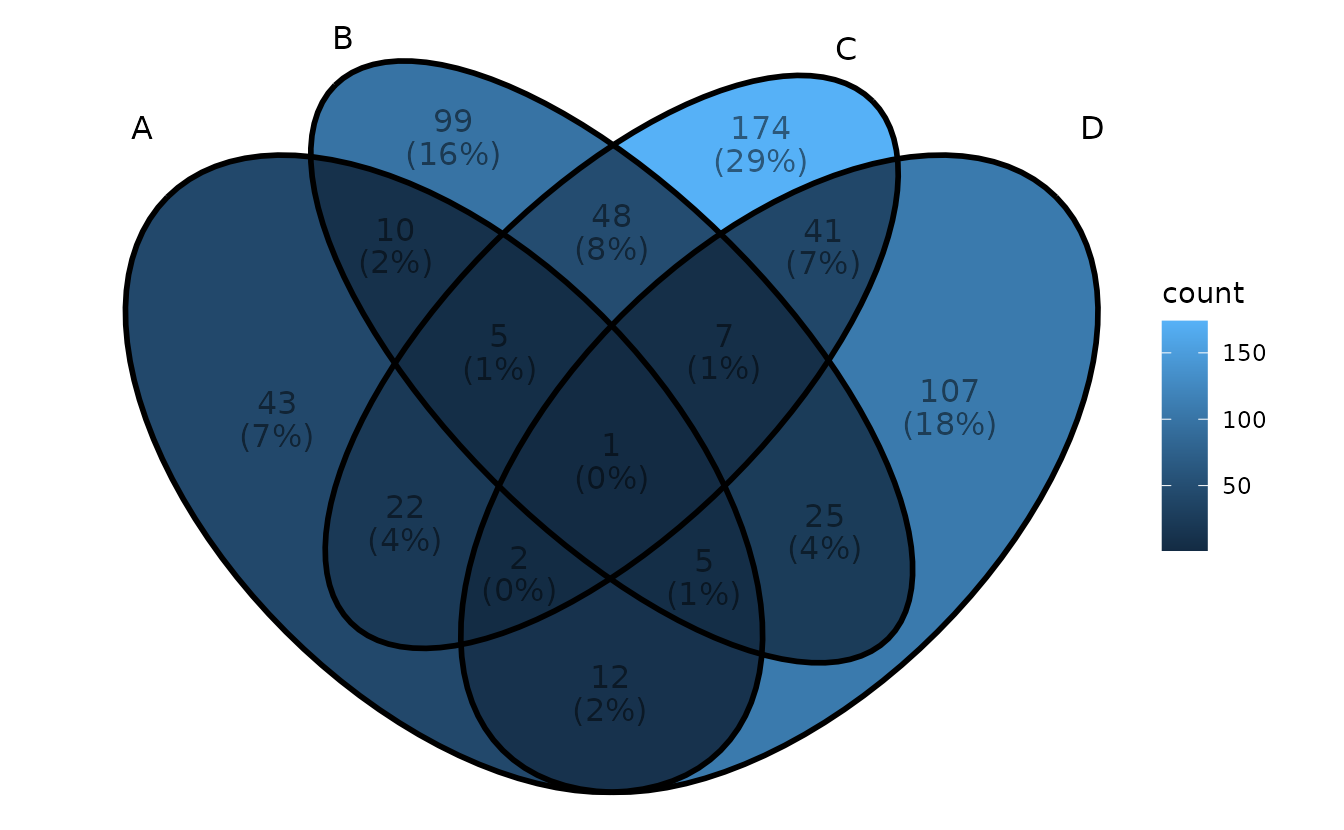
Changing palette
- changing fill palette
library(ggplot2)
p <- ggVennDiagram(gene_list)
# Red Blue
p + scale_fill_distiller(palette = "RdBu")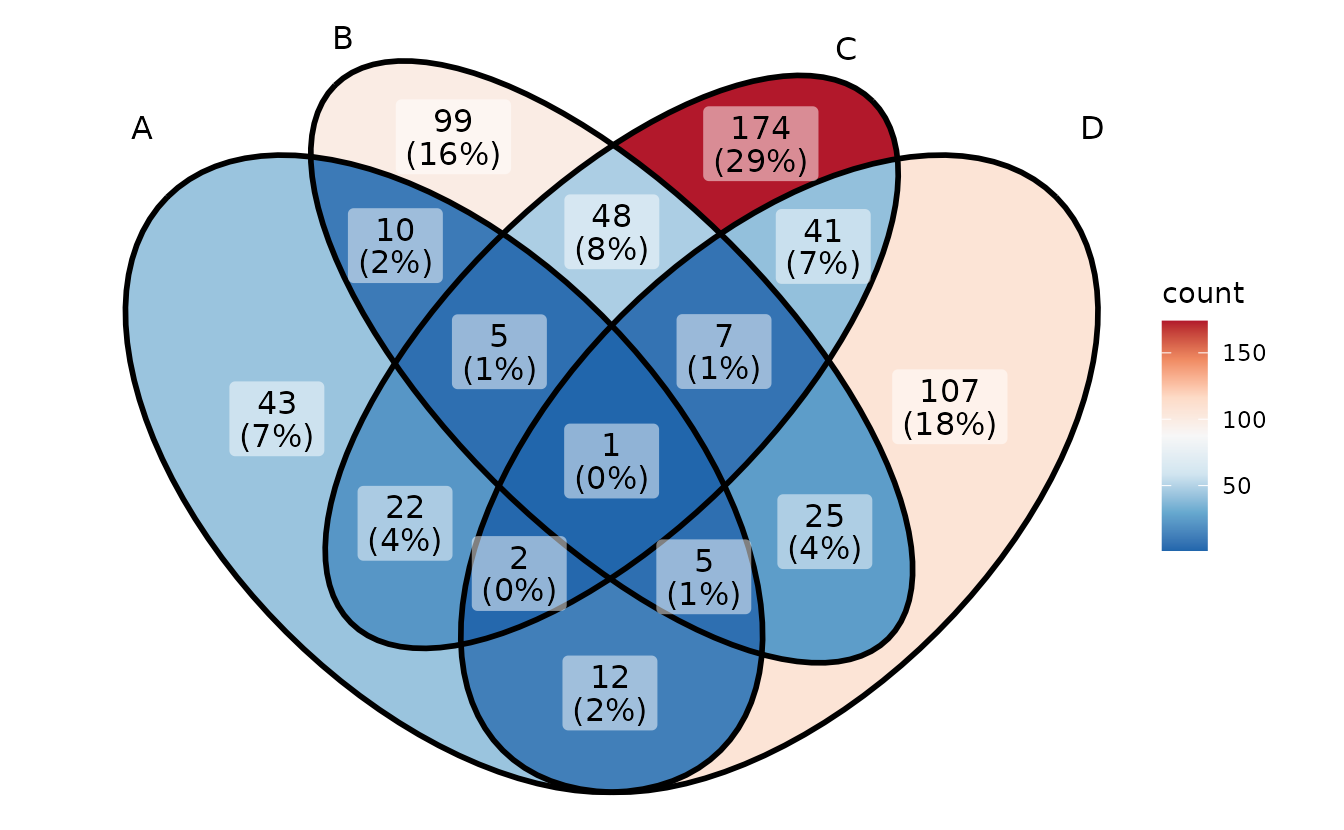
# Reds
p + scale_fill_distiller(palette = "Reds", direction = 1)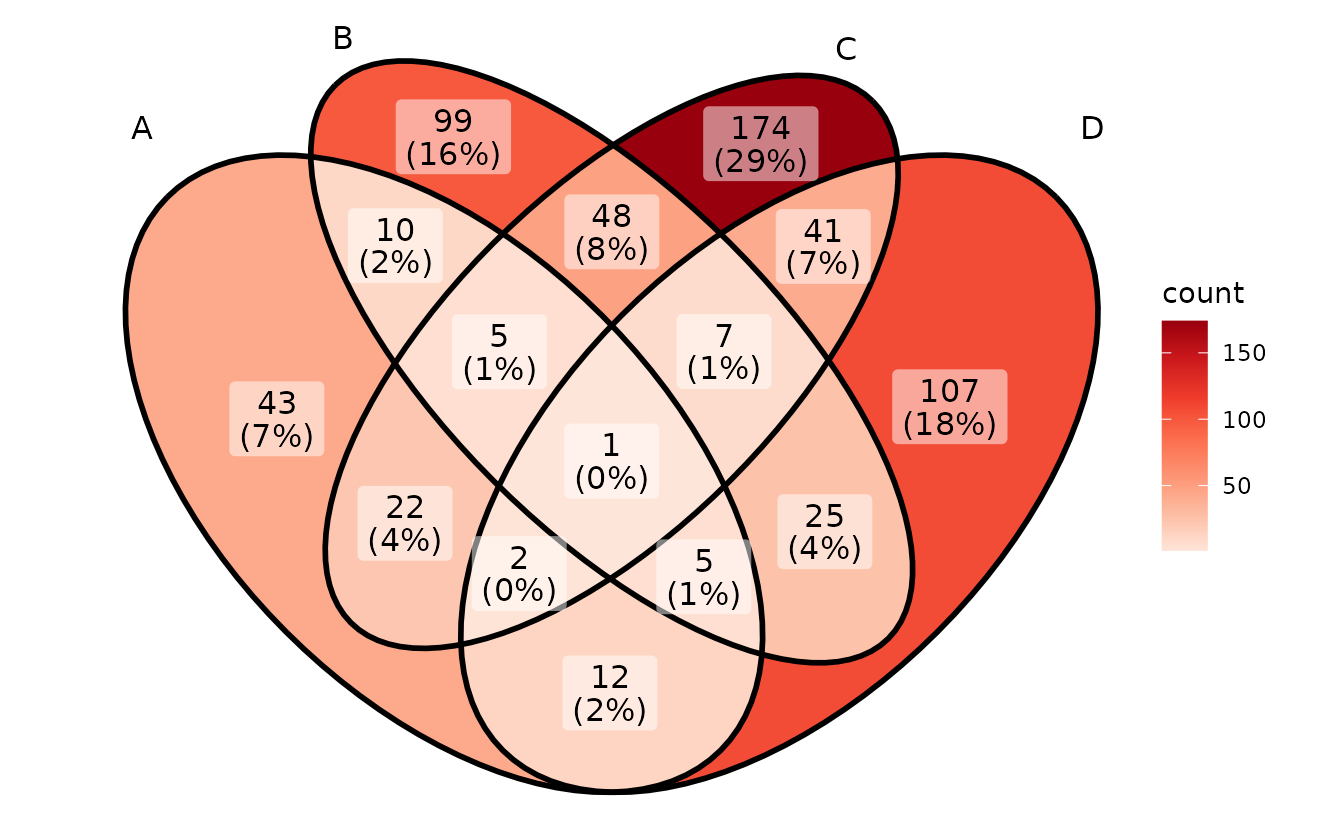
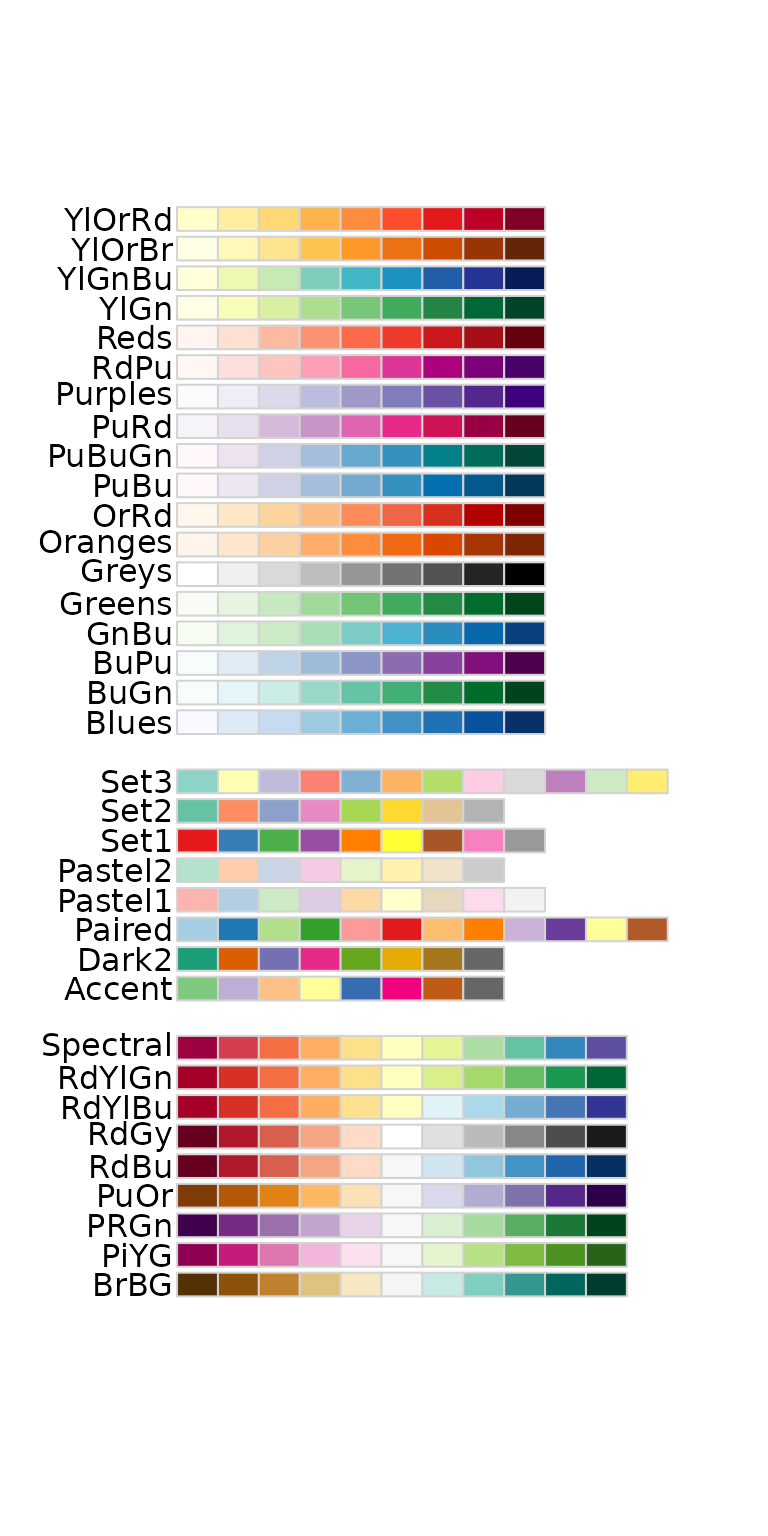
Some other palletes are:
RColorBrewer::display.brewer.all()